| FourierCycle is a tool designed to describe the dynamics of gene-specific mRNA transcription, nuclear export, and degradation rates during the cell cycle. Our biophysical model obtains genome-wide estimates of mRNA transcription, nuclear retention, and degradation half-lives of genes exhibiting oscillatory dynamics in mouse embryonic stem cells. The key feature of single-cell multiome sequencing is that it simultaneously provides readouts for gene expression as well as chromatin accessibility in the same cells. By applying our approach to these data we showcase first-of-its-kind exploration of chromatin accessibility during the cell cycle at high temporal resolution. |
| For more information please read our FourierCycle paper. |
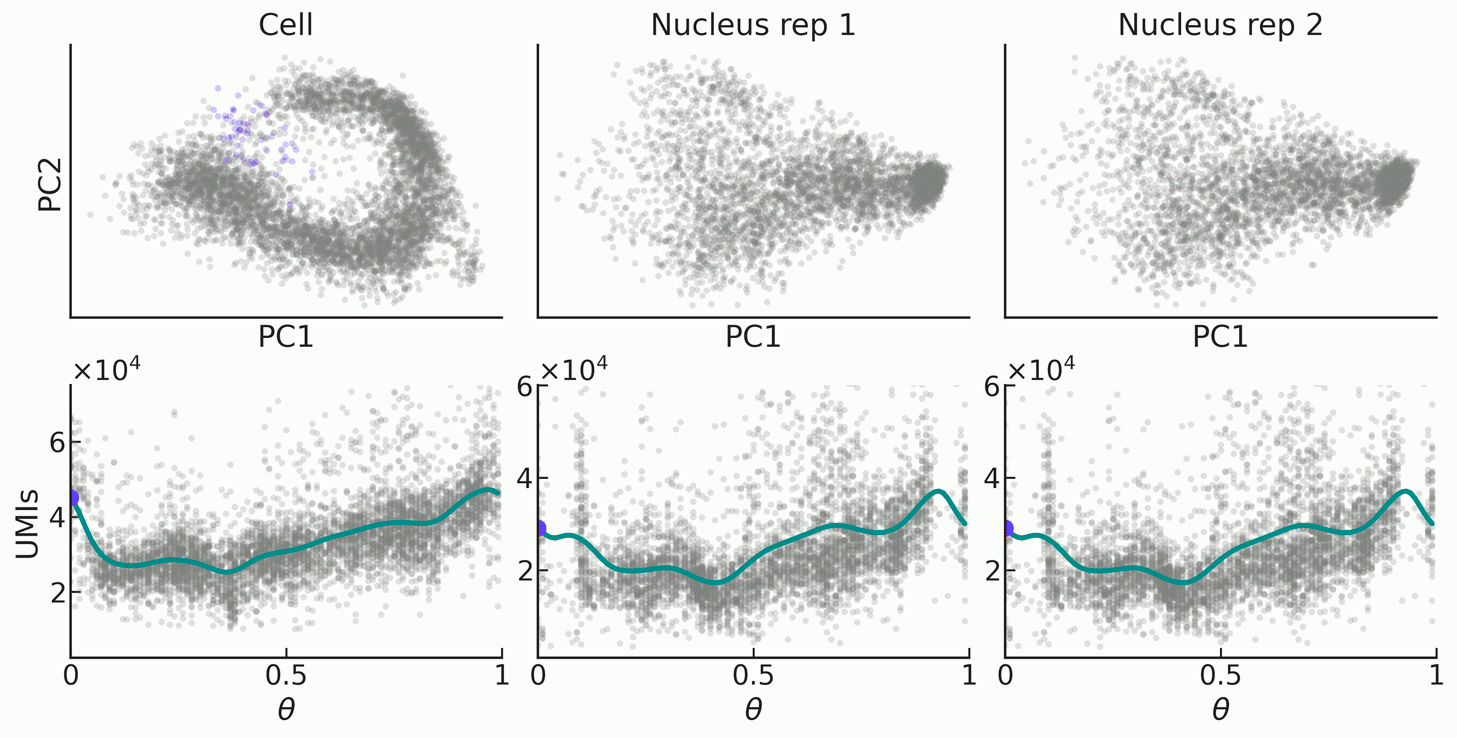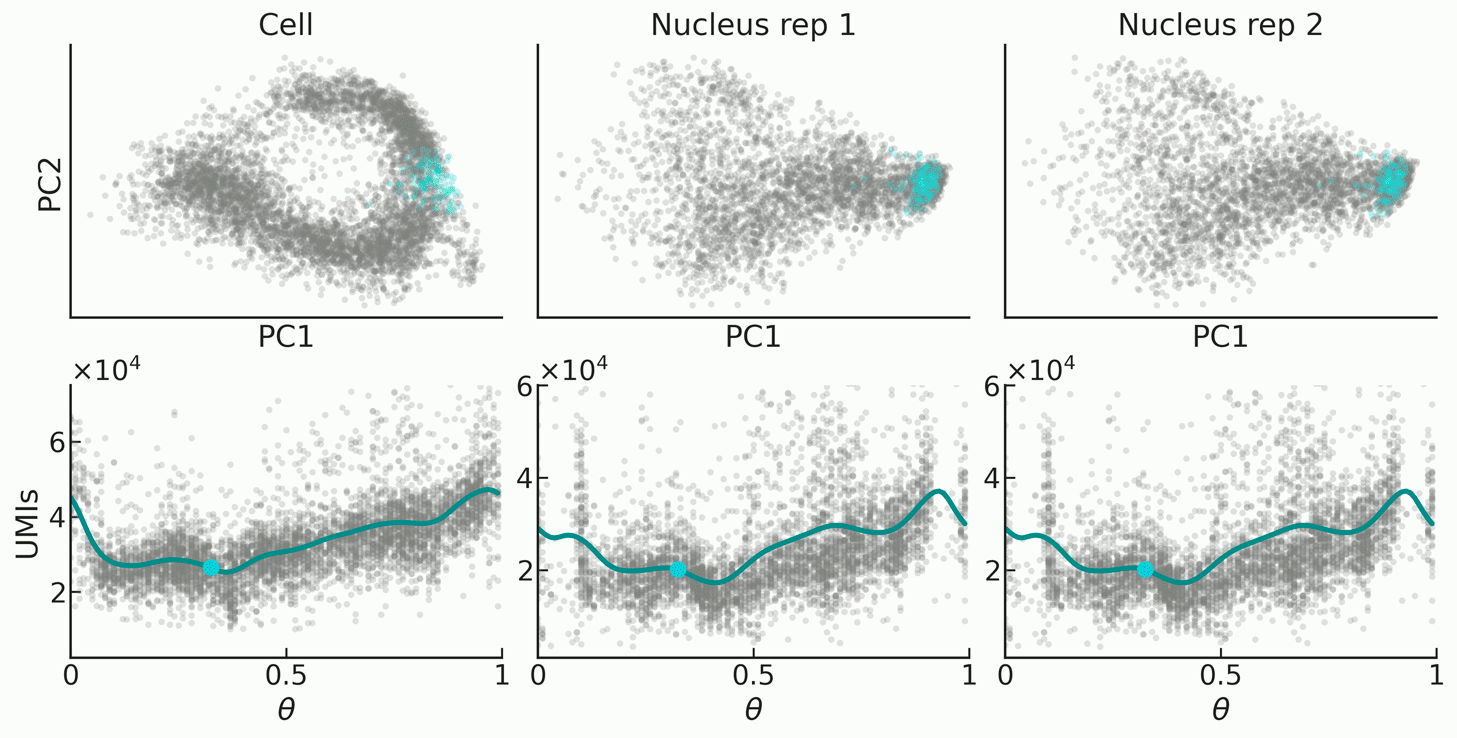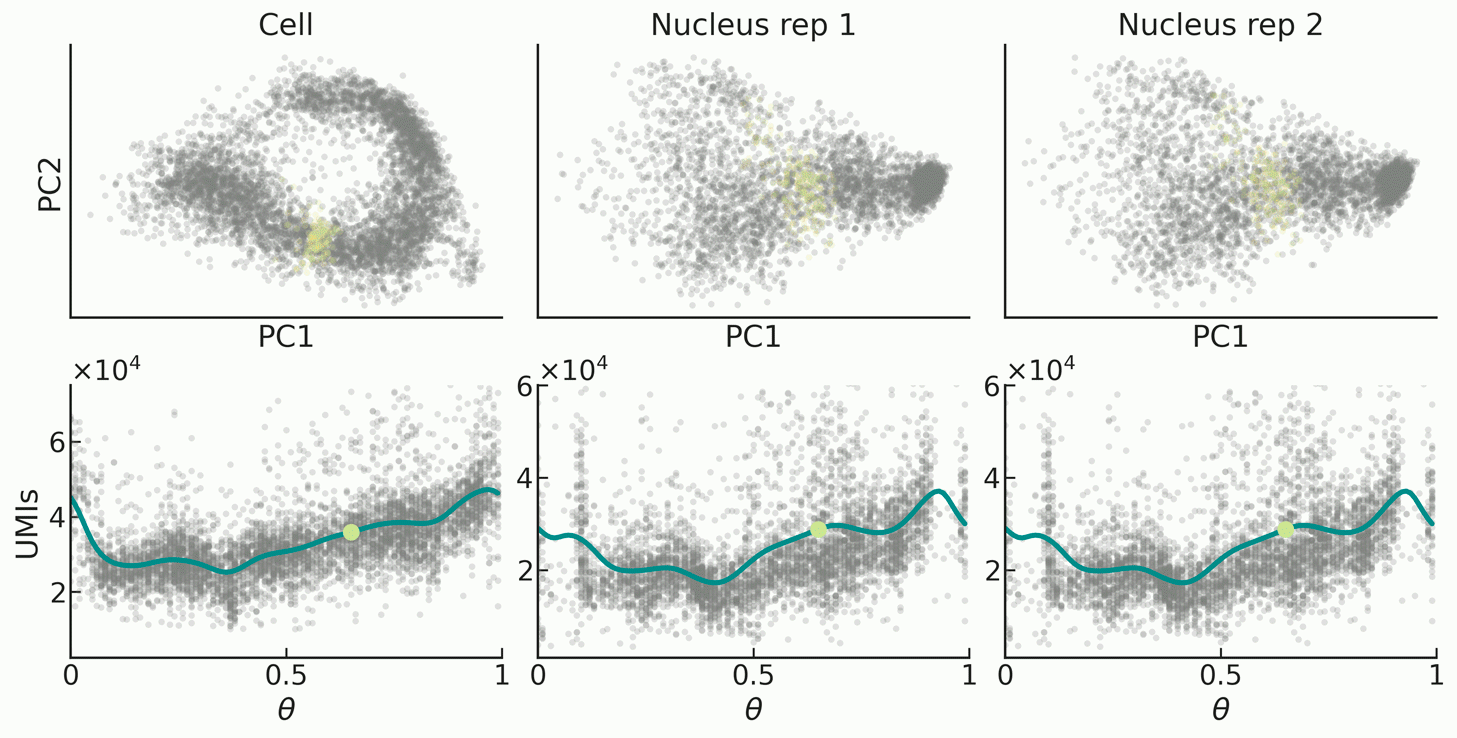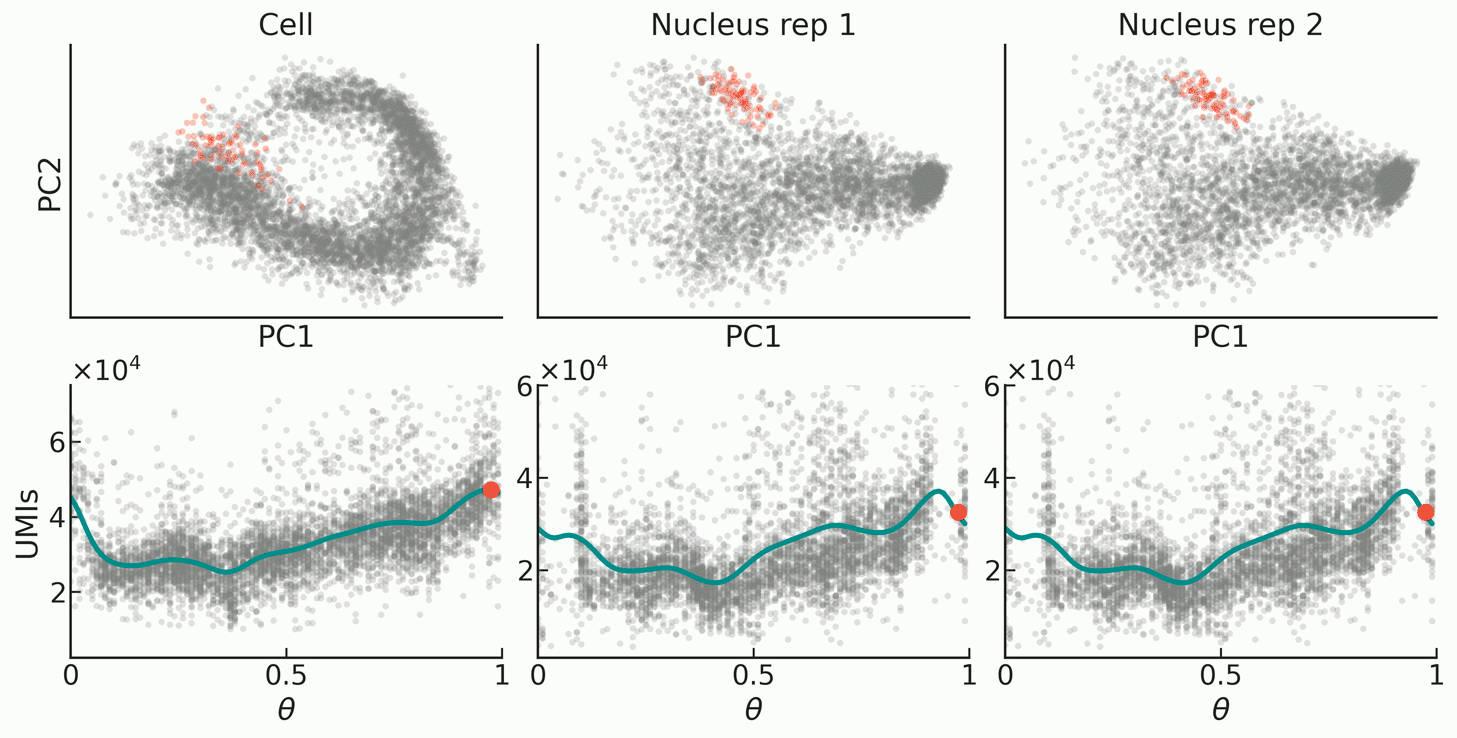 |
